INTRODUCTION
Breast cancer is the most common cancer for women in industrialized countries with the number of breast cancer patients continuously increasing since the 1970s. The etiology of breast cancer is not fully known, and only 5–10% of breast cancer cases can be explained by genetic variation1. Thus, the increasing number of studies indicating that lifestyle, including exposure to toxic chemicals such as dichlorodiphenyltrichloroethane (DDT), perfluorooctane-sulfonamide (PFOSA) and dioxins, can influence the development of breast cancer is highly pertinent to more fully understanding causes of breast cancer2.
More than half of all breast cancers express estrogen receptor alpha (ERα)3. In ER-positive cancers, tumor growth is promoted by the binding of 17-estradiol (E2) to ER3. Since there is an association between elevated levels of estrogen in the blood and higher risk of developing breast cancer, the causes of elevated estrogen levels have been the subject of many studies4. Exposure to endocrine disruptive chemicals (EDCs) with estrogenic activity (‘xenoestrogens’) is thus a recognized factor which can affect hormone balance in mammary cells, potentially increasing overall estrogenic activity in blood and contributing to breast cancer progression5,6. The mammary gland exhibits different sensitivities to estrogen exposure during development. In particular, exposure during the perinatal phase has the potential to dysregulate mammary gland growth and cause cellular transformation, which can be detected later in life5.
Understanding whether chemicals can either initiate or stimulate progression of breast cancer in humans is challenging. The source of exposure to toxic contaminants are multiple and adverse consequences can sometimes be detected only several decades after initial exposure. In order to improve the predictability of toxicity tests, different cell culture model systems have been frequently used to investigate adverse health effects of EDCs. One of the prominent mechanisms investigated by these models is interaction with ER.
MCF-7 is a human breast cancer cell line often used to investigate the effects of EDCs with an estrogenic potential7,8. MCF-7 cells express ERα and their growth is hormone-dependent9. MCF-7 cells can thus provide insight into the mechanisms of hormone-driven breast cancers. In addition, EDCs can sometimes cause effects, which are mediated by mechanisms other than through the ER. Since MCF-7 cells are cancerous and express high levels of ERs, it could be questioned whether this cell model is the most representative of normal breast physiology.
Non-malignant human mammary epithelial cell lines such as MCF-10A are increasingly used as test model systems. Due to their morphological similarity to normal human mammary epithelial cells, MCF-10A cells are widely used in toxicology studies10. MCF-10A cells do not express ERα and are not tumorigenic11,12. MCF-10A cells have been recognized to have many features in common with normal breast epithelium13, but there have also been reports questioning their reliability to represent human breast epithelial cells. MCF-12A is another non-tumorigenic human mammary cell line used to investigate human breast cancer. This cell line has in the past been described as ERα negative. Some studies, however, have reported estrogen-responsive MCF-12A cells14-16 whilst other studies reported weak expression of ER transcripts17 or only identified ERα negative MCF-12A cells18-20. One study investigating the effects of bisphenol A on MCF-12A, concluded that these cells, unlike MCF-10A cells, do express ERα21.
Bisphenol A (BPA) is one of the highest volume-produced EDCs, possibly affecting several endocrine-related pathways6. It is an environmental estrogen used in the synthesis of polycarbonate plastics and resins, which are frequently used in the manufacture of food and drink packaging, baby feed bottles, the coating of DVDs, and thermal receipt and recycled paper. BPA has an analogous molecular structure to estrogen and acts similarly, targeting organs typically regulated by estrogens such as the mammary gland22. There it can bind to ERα, ERβ, and membrane-associated GPR306. Studies concerning adverse health effects due to BPA exposure have caused public concern over its use, and, in response, BPA is now often being replaced by manufacturers with other members of the bisphenol family of compounds. Some of the most commonly used BPA substitutes are bisphenol AF (BPAF), bisphenol F (BPF), and bisphenol S (BPS)23. Many of these alternatives are structurally close to BPA and can thus exert similar estrogenic effects. We previously compared the estrogenic activity of BPA with that of six BPA replacements: BPAF, BPF, BPS, bisphenol AP (BPAP), bisphenol B (BPB) and bisphenol Z (BPZ) in MCF-7 and T47D mammary cell lines; and showed that many of these BPA analogues were as potent or more potent than BPA. BPAF demonstrated the greatest estrogenic capability followed by BPB >BPZ >BPA >BPF >BPAP >BPS8. The results of our study showed that bisphenol alternatives used in ‘BPA-free’ products can often be even more estrogenic and thus more toxic than BPA.
In this follow-up study, we have investigated the effects of BPA alone and a mixture of 7 bisphenols (BPA, BPAF, BPB, BPZ, BPF, BPAP, BPS) on three different human mammary epithelial cell lines (MCF-7, MCF-10A, MCF-12A) at a cell proliferation and transcriptome level.
METHODS
Chemical test substances and other reagents
BPA, BPAF, BPAP, BPB, BPF, BPS and BPZ were purchased from Merck KGaA (Sigma-Aldrich, Gillingham, Dorset, UK). Stock solutions of the test substances were individually prepared in dimethyl sulphoxide (DMSO). Unless otherwise specified, all other reagents and chemicals were purchased from Sigma-Aldrich (UK).
Cell tissue culture and treatments
MCF-7 and MCF-10A cell lines were purchased from the ECACC General Cell Collection (UK Health Security Agency, Porton Down, Salisbury, UK). MCF-12A cells were a gift from E. Silva (Brunel University of London). MCF-7 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM; ThermoFisher Scientific, Loughborough, UK) supplemented with 2 mM glutamine (GE Healthcare Life Sciences, Chalfont St Giles, UK), 10 μg/mL penicillin/streptomycin (ThermoFisher Scientific) and 5% fetal bovine serum (FBS, Merck). MCF-10A and MCF-12A cells were cultured in DMEM-F12 (ThermoFisher Scientific) supplemented with 20 ng/mL epidermal growth factor (EGF), 0.5 μg/mL hydrocortisone (ThermoFisher Scientific), 100 ng/mL cholera toxin, 10 μg/mL insulin (ThermoFisher Scientific), 10 μg/mL penicillin/streptomycin (ThermoFisher Scientific) and 5% horse serum (ThermoFisher Scientific). All cell cultures were maintained at 37℃ in a 5% CO2 – air atmosphere, in 75 cm2 flasks (Corning, Tewksbury, USA).
Treatment of tissue culture cells with bisphenols was at the following concentrations: BPA-high consisted of a high dose of BPA at 250 nM, which is known to impart estrogenic effects8; a lower dose of BPA designated as BPA-low (50 nM), which does not produce proliferative effects in MCF-7 cells8 was also tested; we used the whole-mixture approach to test three mixtures of bisphenols. The mixture designated as BPx-high consisted of 50 nM each of BPA, BPAF, BPAP, BPB, BPF, BPS and BPZ. The same 50 nM BPx-high mixture of bisphenols but lacking BPA (BPx-high-noBPA) was also tested to evaluate whether the bisphenols have effects in the absence of BPA. A mixture of bisphenols at low doses termed BPx-low corresponded to 1 nM each of BPA, BPAF, BPAP, BPB, BPF, BPS and BPZ.
Cell proliferation assays
The three cell lines were each seeded in 32 wells of 48-well plates (Corning, Tewksbury, USA) at a density of 8×103 cells/mL in a volume of 250 μL. After 24 hours incubation to ensure cell attachment, cells were exposed to the different concentrations of the test substances for 6 days. Avoidance of interference from estrogenic substances present in the sera added to the culture media, was accomplished by employing steroid hormone stripped FBS (ThermoFisher Scientific). Horse serum was similarly stripped of its estrogenic substances by treatment with dextran-coated charcoal as previously described24. The dilutions of test substances were refreshed every three days. Each concentration of test substances was assessed in quadruplicate. As a negative control, media containing stripped sera were used. After six days, an MTT assay was conducted to evaluate cell proliferation and viability as previously described8. Briefly, cells were incubated for 2 hours in 100 μL of MTT solution, consisting of MTT and phenol-free DMEM at a concentration of 1 mg/mL. After the incubation, the cells were lysed with DMSO and proliferation determined by optical density measurement of the insoluble formazan produced using a microplate reader (GloMax® Multi Microplate Multimode Reader, Promega, Madison, USA).
RNA extraction
A 48-well plate was seeded as described for the proliferation assays and after 24 hours the cells were exposed to the test compounds. After 48 hours exposure to the test substances, total RNA was extracted using the Qiagen RNeasy Mini Kit (Qiagen, Crawley, UK). RNA concentrations were measured using a NanoDrop instrument (NanoDrop™ One Microvolume UV-Vis Spectrophotometer; ThermoFisher Scientific). This was followed by an assessment of RNA integrity by 1.5% w/v agarose gel electrophoresis (data not shown). RNA samples were then subjected to full transcriptome analysis under contract with GENEWIZ (Leipzig, Germany).
RNA Library Preparation and NovaSeq Sequencing
RNA Library Preparation and cDNA Sequencing were performed by GENEWIZ (Germany). RNA samples were quantified using a Qubit 4.0 Fluorometer (ThermoFisher Scientific) and RNA integrity was checked with RNA Kit on an Agilent 5300 Fragment Analyzer (Agilent Technologies, Palo Alto, CA, USA). RNA sequencing library preparation employed the NEBNext Ultra II RNA Library Prep Kit for Illumina following the manufacturer’s instructions (NEB, Ipswich, MA, USA). Briefly, mRNA was first enriched with oligo(dT) beads and then fragmented. First strand and second strand cDNA were subsequently synthesized. The cDNA fragments were end-repaired and adenylated at their 3’ends. Universal adapters were then ligated to the cDNA fragments followed by index addition and library enrichment with limited cycle PCR. Sequencing libraries were validated using the NGS Kit on the Agilent 5300 Fragment Analyzer (Agilent Technologies) and quantified by using the Qubit 4.0 Fluorometer (ThermoFisher Scientific). The sequencing libraries were multiplexed and loaded on the flow cell of the Illumina NovaSeq 6000 instrument according to the manufacturer’s instructions. The samples were sequenced using a 2×150 Pair-End (PE) configuration v1.5. Image analysis and base calling were conducted by the NovaSeq Control Software v1.7 on the NovaSeq instrument. Raw sequence data (.bcl files) generated from Illumina NovaSeq were converted into fastq files and de-multiplexed using Illumina bcl2fastq program version 2.20. One mismatch was allowed for index sequence identification.
Statistical analysis
The data collected from the cell proliferation assays were analyzed using GraphPad software (GraphPad Software, Inc., La Jolla, California, USA). Data were analyzed using a 5-parameter logistic regression, while the other cell proliferation data were analyzed using a one-way ANOVA. The raw RNA-seq data received from GENEWIZ were analyzed using FASTQC Version 0.11.9. The statistical analysis of the RNA-seq data was done in R studio (R Core Team, 2021). RNA-seq data were first processed with Salmon25. This tool was used to quantify transcript abundance by mapping the reads against a reference transcriptome (Homo sapiens GRCh38 cDNA fasta). The Salmon output was then imported in R using the Bioconductor package tximport. A transcript database containing transcript counts was created, which was used to perform a differential gene expression analysis using DESeq226. To identify genes that were significantly differently expressed, a minimum fold-change of 1.5 and a p<0.05 was used. Next, a KEGG pathway enrichment analysis was conducted using the goseq R Bioconductor package in R studio. All the raw data are available in a publicly available repository (NCBI GSE182963).
RESULTS
The aim of this study was to evaluate action of different bisphenol plasticizers at a cell proliferation and transcriptome level with a particular focus on whether these substances could alter cell function in the direction of carcinogenesis. Experiments were conducted on two non-cancerous primary human mammary epithelial cell lines, MCF-10A and MCF-12A, and one cancerous human mammary epithelial cell line, MCF-7.
The following concentrations of test compounds was employed. BPA-high, consisting of 250 nM BPA, which is known to impart estrogenic effects8 and BPA-low (50 nM BPA), which does not produce proliferative effects in MCF-7 cells8. We used the whole-mixture approach to test three mixtures of bisphenols.
Effects of BPA and BPA analogue mixtures on mammary epithelial cell proliferation and viability
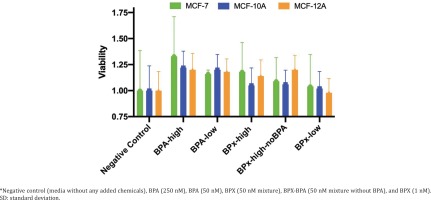
We first undertook cell proliferation and viability assays to determine at which concentrations the test compounds could exert estrogenic activity and cytotoxicity. Overall, the varying concentrations of the test substances (BPA-low, BPA-high, BPx-high, BPx-high-noBPA, BPx-low) showed a limited capability to stimulate growth of the three mammary epithelial cell lines (MCF-7, MCF-10A, MCF-12A) (Figure 1). Nevertheless, a trend was observed in that all three cell lines showed a higher proliferative response after the exposure to the BPA-high and BPA-low concentrations as well as BPx-high and BPx-high-no mixtures. No increase in cell proliferation was observed with the BPx-low mixture.
Transcriptome analysis
Since steroid hormone stripped horse serum was not commercially available, we prepared both horse and FBS stripped sera using the same protocol (see Methods section). This was used in cell cultures for subsequent transcriptome analysis, knowing that these self-stripped sera can maintain good cell viability (Figure 1). Samples of MCF-7, MCF-10A and MCF-12A cells were thus cultured in a medium containing self-stripped sera and treated with BPA-high (250 nM), BPA-low (50 nM), BPx-high (50 nM bisphenol mixture), BPx-high-noBPA (50 nM bisphenol mixture without BPA) and BPx-low (1 nM bisphenol mixture) test substances, for 48 hours before total RNA extraction for transcriptome analysis.
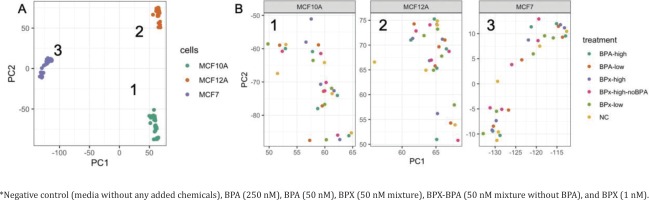
Raw RNA sequencing data were analyzed to identify differentially expressed genes (DEG) and affected pathways. Initially, a principal component analysis (PCA) was conducted to identify the main sources of variation in gene expression profiles (Figure 2 A). The first 2 components were mainly driven by the separation between the three cell types. The cancerous cell line MCF-7 was separated from the two primary non-cancerous cell lines MCF-10A and MCF-12A along the first component, while the transcriptome from MCF-10A and MCF-12A was separated along the second component. This indicated that MCF-10A and MCF-12A are relatively similar and different from MCF-7. A further evaluation of these three clusters (Figure 2 B) showed no full clear separation according to the treatment groups.
Figure 2
Principal component analysis reveals the sources of variations in the comparison of gene expression profiles after exposure* to bisphenols of MCF-7, MCF-10A, MCF-12A: A) General PCA showing the clustering of the samples from the three cell lines along the PC1 and PC2 axes. B) Focus on the treatments for the gene expression profiles extracted from MCF10A (cluster 1), MCF12A (cluster 2) and MCF7 (cluster 3)
Analysis of the number of DEG revealed that the three cell lines displayed different responses to the bisphenol treatments (Table 1). MCF-10A was the most affected with a higher number of upregulated and downregulated genes compared to the MCF-7 and MCF-12A cell lines. It is noteworthy that there is no clear increase in the number of genes affected by BPA alone when comparing the high (250 nM) and low (50 nM) concentration exposures (Table 1). We next investigated whether identical DEG are present across the different cell lines (Table 2). This analysis showed that there was no overlap of affected upregulated genes. However, some downregulated genes were affected by the same treatment in two out of the three cell lines. Firstly, VASN coding for the protein Vasorin was similarly affected in MCF-7 and MCF-12A cells after exposure to BPA-high (250 nM) and BPx-low (1 nM) concentration mixture. Secondly, DENND11 (DENN Domain Containing 11) was downregulated in MCF-7 and MCF-10A cells after being treated by BPA-low (50 nM), and the BPx-high (50 nM) and BPx-low (1 nM) bisphenol mixtures. Additionally, CORO7, which encodes the Coronin 7, was downregulated in the MCF-7 and MCF-12 cell lines after exposure to the BPx-high-noBPA (50 nm) mixture.
Table 1
Differentially expressed genes (DEG) from transcriptome analysis of MCF-7, MCF-10A and MCF-12A cell lines after 24-hour exposure to treatments*
Table 2
Genes that were differentially expressed in more than one of three mammary epithelial cell lines (MCF-7, MCF-10A and MCF-12A) after exposure to the same treatment
| Treatments* | Gene (downregulated) | Gene ID | Cell lines |
|---|---|---|---|
| BPA-high | VASN | ENSG00000168140 | MCF-7, MCF-12A |
| BPA-low | DENND11 | ENSG00000257093 | MCF-7, MCF-10A |
| BPx-high | DENND11 | ENSG00000257093 | MCF-7, MCF-10A |
| BPx-high-noBPA | CORO7 | ENSG00000262246 | MCF-7, MCF-12A |
| BPx-low | VASN | ENSG00000168140 | MCF-7, MCF-12A |
| DENND11 | ENSG00000257093 | MCF-7, MCF-10A |
As the number of identical DEG across different treatments was too large to investigate on an individual gene basis, we undertook a KEGG pathway enrichment analysis to determine which pathways are most affected by the observed alterations in gene expression profiles. However, only the DEG data from MCF-10A cells could be subjected to this analysis as the number of DEG for MCF-7 and MCF-12A was too low to allow such an investigation. Table 3 summarizes the most affected pathways in MCF-10A cells from each treatment. Pathways and molecular relation networks such as oxidative phosphorylation, spliceosome, and pathways in cancer, were affected after exposure to different treatments. The ubiquitin-mediated proteolysis pathway and metabolic pathways were the most dysregulated, as they were affected after exposure to all treatments. Altogether, these results show that while limited effects were detected measuring cell growth, the evaluation of gene expression profiles was more sensitive with the exposure of MCF-10A cells to bisphenols, potentially revealing new mechanisms of toxicity for these compounds.
Table 3
The most affected pathways overrepresented in MCF-10A predicted from transcriptome analysis§
| Pathway | BPA-high | BPA-low | BPx-high | BPx-high-noBPA | BPx-low |
|---|---|---|---|---|---|
| Metabolic pathways | *** | ** | *** | *** | ** |
| Protein processing in ER | *** | ** | ** | *** | * |
| Spliceosome | ** | * | ** | ** | |
| Ubiquitin mediated proteolysis | * | ** | ** | ** | * |
| Ribosome | * | * | * | ||
| Oxidative phosphorylation | * | * | |||
| Parkinson disease | * | * | |||
| Nucleocytoplasmic transport | * | * | |||
| Lysosome | * | ||||
| Endocytosis | * | ** | * | ||
| Huntington disease | * | ** | |||
| mRNA surveillance pathway | * | ||||
| Autophagy - animal | * | * | |||
| Pathways in cancer | ** |
§ MCF-10A were exposed to BPA-high (high concentration; 250 nM), BPA-low (low concentration; 50 nM), BPx-high (50 nM bisphenols mixture), BPx-high-noBPA (50 nm bisphenols mixture excluding BPA), and BPx-low (1 nM bisphenols mixture), for 24 hours. Total RNA was extracted and subjected to transcriptome analysis. The identification of differentially expressed genes led to KEGG pathway enrichment analysis to determine most affected pathways. Statistical analysis was enrichment in KEGG pathway.
DISCUSSION
Bisphenols are found in many everyday items, such as plastic drinking bottles containing carbonated drinks, the resin lining cans containing food and drink, and as a covering on thermal receipt paper. Thus, bisphenols can readily be absorbed into the human body on a daily basis. As bisphenols have a proven estrogen mimicking capability, this can lead to dysregulation of the endocrine system and can increase the risk of developing cancers, especially breast cancer6. BPA is one of the most highly manufactured xenoestrogens with a large body of evidence linking this compound with a higher risk of developing breast cancer. Within the European Union, proposals have been tabled by the European Food Safety Authority to slash BPA intake in adults by 105-fold from 4 μg to 0.04 ng per kg body weight per day27. In response to public concerns and proposed regulatory changes, industry is increasingly replacing BPA with other bisphenol family members and marketing its products as ‘BPA-free’. However, these bisphenol substitutes often bear a similar or greater potential to disrupt the balance of the endocrine system being more estrogenic than BPA8 leading to the possibility that in many instances this could be a case of ‘regrettable substitution’. However, the exact mechanisms by which bisphenols exert their toxic effects and their consequences, is not fully understood. In this study, we describe differences in the response to bisphenols at a gene expression level between three mammary cell lines.
Analysis of DEG across the three cell lines revealed that MCF-10A was more affected by exposure to the test substances. This comes as somewhat of a surprise, as out of the cell lines employed, MCF-10A is the only one which does not express ERα11,12. Nevertheless, based on previous studies it is known that bisphenols can also affect cell function via estrogen receptor independent pathways28, which is most likely the case in MCF-10A. A study on the proliferative effects of low concentrations of BPA on ERα-negative cell lines, including MCF-10A, has shown that it can lead to upregulation of oncogenic cell-cycle regulatory proteins resulting in proliferation and DNA damage29.
The expression of only three genes was commonly affected across more than one cell line, which was expected as the number of DEG in MCF-7 and MCF-12A cells was low. VASN was downregulated in MCF-7 and MCF-12A cells after exposure to BPA-high (250 nM) concentration and to BPx-high (50 nM) bisphenol mixture. VASN is a highly conserved gene, which codes the protein Vasorin, which regulates TGF-β signaling30. It is highly expressed in different cancer cell lines and can lead to tumor progression31-33, and in gliomas, for example, its expression correlates with tumor aggressiveness33. However, the downregulation of VASN expression observed here implies that it does not play a role in breast cancer. DENND11 was downregulated in MCF-7 and MCF-10A cells following exposure to BPA-low (50 nm) as well as the BPx-high (50 nm) and BPx-low (1 nm) bisphenol mixtures. DENND11 encodes for the protein DENN domain-containing protein 11, which is known to interact with Rab GTPases34. The third downregulated gene across at least two cell lines was CORO7, which encodes the protein coronin 7, localized in the Golgi complex, and is involved in the structure and function of the Golgi35. No pathway connection was found between any of these three genes.
As only a low number of DEG were found in the MCF-7 and MCF-12A cell lines, the focus of the pathway analysis was on MCF-10A. The most affected pathways across treatments were metabolic pathways and the ubiquitin-proteasome pathway (UPP). The UPP is one of two identified intracellular protein degradation pathways and is responsible for the majority of cellular protein degradation, which is essential to maintain a dynamic balance between protein biosynthesis and degradation36. Ubiquitin-mediated degradation is especially important in cell-cycle control, as it controls the number of regulatory molecules present in cells. Dysregulation of the UPP can cause alterations in the cell cycle and can consequently lead to tumor development36. The function of the spliceosome, which is another affected pathway in this study, is also known to be involved in the etiology of breast cancer. For instance, the expression of spliceosome-related genes is known to be pivotal for breast cancer survival37.
Several reports have established a mutual regulation between ERs and the UPP. ERα is degraded by the UPP in a ligand-dependent manner38 and inhibiting polyubiquitination of ERα leads to a decrease in its degradation and to higher steady-state levels of ERα in cells39. Furthermore, it has been shown that activation of the ubiquitin-proteasome system reduces the levels of ERα in human endometrial cancer cells40. While the MCF-10A cell line does not express ERα, it does express G protein-coupled estrogen receptor 1 (GPER)41. A study conducted on MCF-10A cells showed that E2 can lead to GPER-dependent proliferation42. Furthermore, GPER may also be a predictor of aggressive breast cancer types as its high expression often correlates with increased tumor size and the presence of distant metastasis42. The UPP is one of the two ways by which GPER is degraded as it can be ubiquitinated, transported to the Golgi complex, and further phosphorylated before proteolysis39. Therefore, dysregulation of the UPP in MCF-10A cells can lead to an imbalance of degradation and synthesis of proteins controlled by the UPP, such as GPER, which can consequently lead to higher levels of estrogen receptors in general being present in cells and thus culminating in pathological cellular events. The same pathway could also contribute to breast cancer risk in MCF-7 and MCF-12A cells, by not only increasing the levels of GPER but also ERα. As the number of DEG was not sufficient for a pathway analysis in MCF-7 and MCF-12A, dysregulation of the UPP in these cell lines could not be established but has been the subject of other studies43,44. A previous investigation by Xia et al.44, for example, has shown that ubiquitin specific protease 7 (USP7) interacts with ERα and mediates deubiquitylation and stabilization of this receptor. Consequently, the use of an USP7 inhibitor led to increased apoptosis of ERα-positive tumor cells44. Some studies have shown that estrogenic activation of the UPP is linked to cell proliferation and poor survival in late-stage ERα-positive breast cancers45. It thus suggests that alterations in protein processing in the ER, in particular the disturbance of the UPP pathway, could be a mechanism by which exposure to bisphenols could be linked to cancer initiation and progression.
Most studies on bisphenols focus only on one member of this family of compounds and do not explore synergistic, additive or antagonistic effects that these substances can have on the human body resulting from co-exposure. Therefore, future research needs to evaluate the mechanisms of multiple environmental toxins acting simultaneously. Additionally, as the general population is not only exposed to one specific class of compounds, a mixture of endocrine-disrupting chemicals of different classes should be tested. Another knowledge gap is the exact mechanisms of action and toxic effects of BPA alternatives on the reproductive system. Prenatal exposure to BPA is associated with morphological and functional alteration in female reproductive organs, such as ovaries, oocytes and the mammary gland, as well as male reproductive organs46. Estrogenic or anti-androgenic effects of BPA alternatives could lead to similar effects.
This study also underlines the importance of using different cell model systems. Another aspect that should be considered when choosing cell models is ethnic background and sex. Primary mammary epithelial cell lines of different ethnic backgrounds could help to investigate possible varying susceptibilities between ethnic groups47. Additionally, the use of cell models from male individuals could help to evaluate the breast cancer risk these endocrine-disrupting chemicals have in men. Generally, the breast cancer risk for men is often underestimated and should be considered more often in studies48. Furthermore, endocrine disruptors sometimes do not have a linear dose-response and testing a large range of concentrations could offer insight into different dose responses49. Finally, biomonitoring of a human population could help to evaluate the levels and different mixtures of compounds to which human populations are exposed.
Further research needs to be conducted in animal model systems. Some studies have already reported the contribution of BPF and BPS in the etiology of breast cancer by showing pro-tumorigenic changes in primary human mammary gland organoids50. An investigation in a human breast cancer xenograft model system showed that BPS promoted breast cancer cell proliferation with a non-monotonic dose response51. Some studies in humans are beginning to provide indications that BPA concentrations in the breast tissue of cancerous patients are significantly higher than in non-cancerous tissue52.
CONCLUSIONS
This study shows that different breast cancer cell model systems are differently affected by exposure to BPA and mixtures of bisphenols used as BPA replacements. This demonstrates the value of using multiple mammary epithelial cell types to obtain a more accurate toxicity profile stemming from exposure to this class of chemicals. Our results show that the evaluation of the transcriptome profile of MCF-10A cells treated with bisphenols reveals potential new mechanisms of toxicity for these compounds. Our results also highlight the value of using ER negative cell lines such as MCF-10A to inform on the potential of endocrine disruption leading alteration in mammary cell function independently of ERα activation.